Snowball Sampling (Labeled Snowball with multiple inclusions)
Source:R/bootstrap-snowboot.R
bootstrap_snowboot.RdThis is a slightly modified version of the lsmi from the snowboot package.
Arguments
- network
An igraph object with \(n\) nodes.
- B
Number of bootstrap samples
- num.seed
Number of seeds to be included each the snowball sample
- num.wave
Number of waves to be included each the snowball sample
- output.type
The type of object the resulting bootstrap networks should be. The default is
igraphwhich will make bootstrap samples of class "igraph". Note that for large \(B\), this may not be an efficient use of storage space. Other options includeedgelistwhich returns an edge list for each bootstrap sample,matrixwhich returns bootstrap samples as an \(n\)-by-\(n\) adjacency matrix,dgCMatrixwhich returns sparse matrices (packageMatrixmust be loaded). See make_network_type for details.
Value
A list of length \(B\) where each element is an bootstrap sample.
Each element is of class output.type.
Details
This is a slightly modified version of the lsmi from the snowboot package.
This function inputs a igraph object, it uses the node names (not node IDs),
and it returns the edge list of the bootstrap sample (not the nodes included in the sample).
Most code in this function is directly from lsmi from the snowboot package. This function is designed to be a wrapper function to format the lsmi results into the JaB package syntax.
References
Ramirez-Ramirez L, Nezafati K, Chen Y, Lyubchich Y, Gel YR (2022). snowboot: Bootstrap Methods for Network Inference. R package version 1.0.2, https://cran.r-project.org/web/packages/snowboot.
Gel YR, Lyubchich V, Ramirez LR (2017). “Bootstrap quantification of estimation uncertainties in network degree distributions.” Scientific Reports, 7(1). doi:10.1038/s41598-017-05885-x .
Examples
library(igraph)
data("karate")
# Snowboot Bootstrap
set.seed(1)
boot.sample <- bootstrap_snowboot(karate, B = 1, num.seed = 1, num.wave = 2)
#plot comparison of original data and bootstrap sample
par(mfrow = c(1, 2))
#get the same positions in the original data and bootstrap samples
l <- igraph::layout_nicely(karate)
which.index <- NA
for(i in 1:gorder(boot.sample[[1]])){
which.index[i] <- which(V(boot.sample[[1]])$name[i] == V(karate)$name)
}
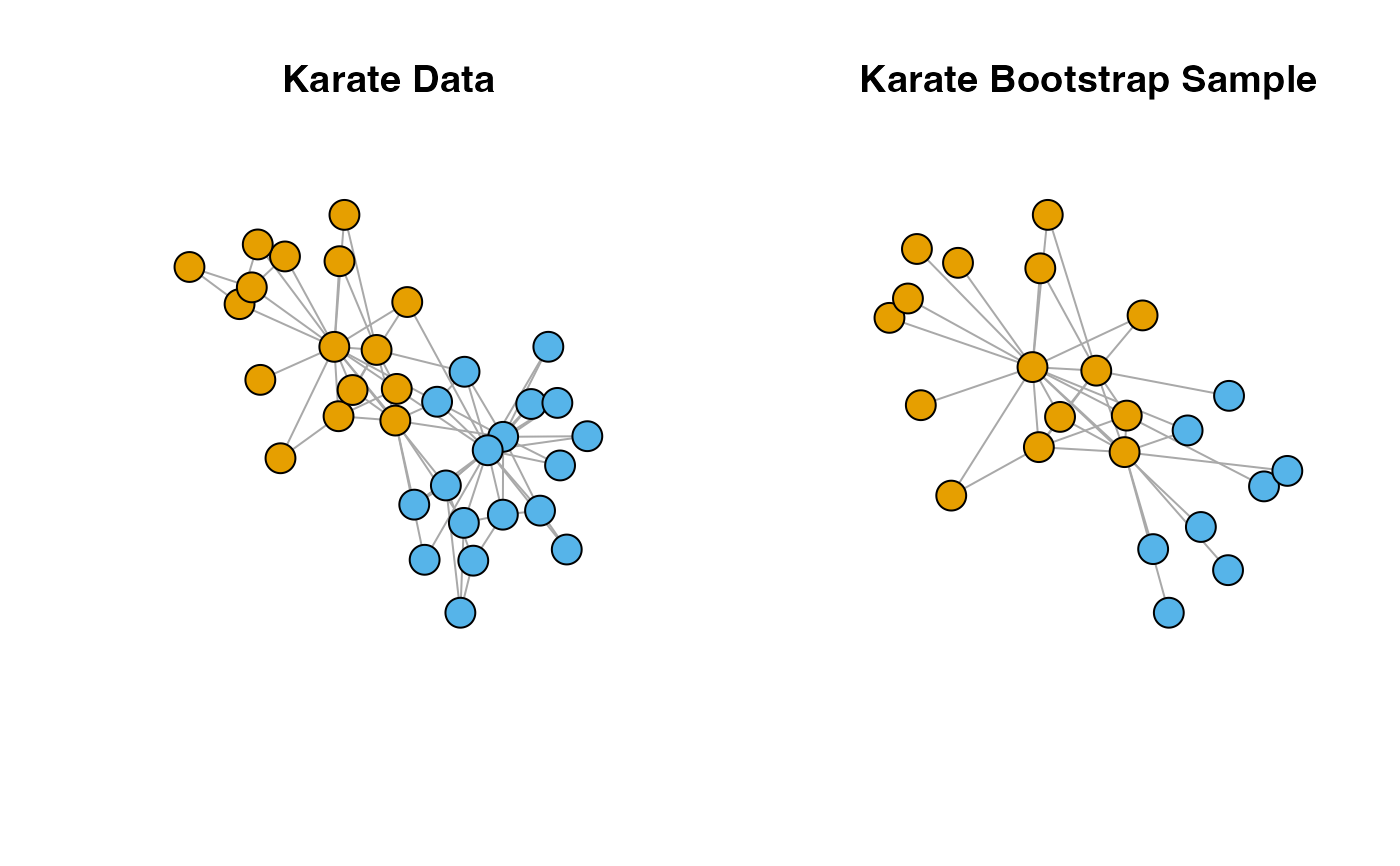
plot(karate,
layout = l,
main = "Karate Data",
vertex.label = NA)
plot(boot.sample[[1]],
layout = l[which.index, ],
main = "Karate Bootstrap Sample",
vertex.label = NA,
vertex.color = V(karate)$Faction[which.index ])